Helicase Structure Solved, Provides Understanding of Molecular Motor
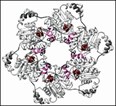

This crystal structure shows six individual helicase proteins assembled into a ring-shaped motor that unwinds the DNA helix. The red balls nestled between two lobes represent dTTP, the motor's fuel molecule, and the pink curl shows the area where the motor grabs onto the DNA.
Helicases, defects of which underlie several human diseases, (cancer-prone Bloom's syndrome and a disease of premature aging called Werner syndrome) work as a molecular motor, threading a strand of DNA through its central hole, ploughing through 300 base pairs per second while shoving the second strand out of the way. Yet, despite its central role in DNA replication, according to Ellenberger, "We know next to nothing about how these helicases move on DNA."
For Ellenberger, the helicase represents a step in his ultimate goal to crystallize the entire replication fork of the bacteriophage T7, a complex of five different proteins that copies DNA. Last year, researchers led by Ellenberger and Charles Richardson, professor of biochemistry and molecular pharmacology, reported the crystal structure of T7's DNA polymerase. Richardson's group showed that the helicase and the other proteins in the replication fork physically touch each other.
The crystal structure of the helicase does not actually represent the way this enzyme occurs in nature. In T7, the helicase exists as a double-decker protein with two enzyme activities. The large helicase donut sits atop a smaller one, the primase, another enzyme of the replication fork. Richardson's lab prepared a helicase fragment of the helicase primase that was amenable to X-ray crystallography. Curiously, this fragment crystallized as an open ring, whereas the biologically active form of the helicase more closely resembles a flat washer. "We think, however, that all interactions we are describing closely approximate what we would see in a closed ring," says Ellenberger.
The researchers found all conserved amino-acid sites near the surface of the donut's hole, where one DNA single strand passes through. More importantly, the structure gives the researchers an inkling of how this motor generates motion from energy.
It was known that the helicase splits a phosphate off dTTP to free up chemical energy. It must somehow convert this into the physical force needed to separate the Watson-Crick bonds joining the double-strand DNA base pairs. It also must harness energy for large conformational changes that allow it to step along the DNA. Finally, it needs a mechanism for grabbing and letting go of DNA.
Ellenberger feels that the crystal structure suggests a plausible scenario as to how the six subunits cooperate to make all this happen. One dTTP is nestled in the cleft between every two subunits, meaning that changes resulting from every dTTP reaction could spread to two subunits.
The dTTP binding site also abuts the DNA binding site at the donut's inner ring. When crystallized without dTTP, these DNA binding sites were disordered and failed to appear as crisp patterns in the crystal structure. Yet when the researchers immersed the helicase crystals in a dTTP solution to allow the dTTP to seep into place, they found that not only was the dTTP sitting in its binding pocket, but the adjoining DNA-binding region of the helicase also suddenly became visible. This suggests that the dTTP reaction might be coupled to DNA binding, in that dTTP cleavage enables the DNA binding region to take shape and grab the DNA. A fraction of a second later, this sequence could occur in the adjacent subunits, and so on throughout the ring.
Other researchers have advanced two major mechanistic models for simpler helicases that are now being hotly debated. Sawaya's structure cannot pick a winner largely because it is symmetrical. It does not visualize the larger conformational changes that must occur within the ring as it advances on the DNA.
The structure does, however, nurture an old passion, says Ellenberger. "As a kid I always loved engines, and I am still fascinated that you can make a protein function as this large, cooperative assembly to move rapidly down a strand of DNA. You've got all these motions flickering in a nanosecond time-realm. This works only because everything is supremely coordinated."
For more information: Tom Ellenberger, Department of Biological Chemistry and Molecular Pharmacology, Harvard University, Cambridge, MA 02138. Tel: 617 432 0458. Fax: 617-738-0516. Email: tome@jeckle.med.harvard.edu.
