Phenotypic HIV testing can predict emergence of resistance
"To date, drug resistance tests have only been used after a patient's treatment fails," said study co-author Steven G. Deeks of the UCSF. "These results indicate that a sensitive phenotypic resistance assay, like the one used in this study, may be useful for predicting and avoiding prolonged treatment failure. I look forward to seeing additional studies on this new frontier in resistance testing."
The assay
PhenoSense HIV directly measures the susceptibility of a patient's virus to reverse transcriptase inhibitors and protease inhibitors. The assay amplifies patient HIV to derive HIV protease (PR) and reverse transcriptase (RT) sequences from a patient plasma sample. A resistance test vector (RTV) is constructed by incorporating the patient-derived segment into a viral vector with an indicator gene, firefly luciferase. Since infected patients typically are infected with mixtures of genetic variants, RTVs are prepared as large pools of sequences, which accurately represent the viral population present in the patient at the time of sample collection.
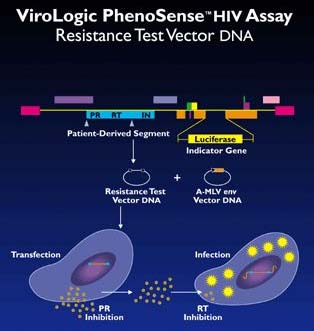
The RTVs are transfected into host cells, virus particles are collected after transfection and used to infect target cells. Serial concentrations of PR inhibitors are added at the transfection step and RT inhibitors at the infection step. Drug susceptibility is measured by comparing luciferase activity produced in the presence and absence of drugs. Susceptible viruses produce low levels of luciferase activity in the presence of PR and/or RT inhibitors, whereas viruses with reduced susceptibility to these drugs produce higher levels of luciferase activity.
For further information on the PhenoSense HIV assay, read Measuring HIV response to antivirals using ViroLogic's PheonSense assay.
The results
The study, conducted by researchers from the UCSF, the Gladstone Institute of Virology, and ViroLogic, retrospectively examined resistance profiles from 16 subjects undergoing second-line antiretroviral therapy. The scientists were able to measure drug susceptibility in samples from patients with viral loads below 100 copies/ml, and detected reduction in drug susceptibility at viral loads as low as 260 copies/ml (for commercial purposes, the PhenoSense HIV assay has been validated for samples with viral loads of 500 copies/ml and above).
In the majority of subjects (six out of ten), the researchers could detect emerging drug resistance prior to significant viral load increases. They conclude that "phenotypic testing has the potential to provide clinical benefit by facilitating earlier change in treatment regimens before the virus develops significant cross-resistance to related drugs." Early use of these highly sensitive drug resistance tests may help preserve effective treatment options, which are often severely limited by drug resistance. The study suggests that drug resistance testing could become a standard monitoring tool to evaluate risk of treatment failure.
Comparing results obtained with phenotypic testing with genotypic resistance testing, an indirect method that uses the presence of mutations to predict resistance, showed that "genotypic analysis... was less valuable in predicting the response to salvage therapy than phenotyping," the researchers report.
"In the future, physicians may use resistance tests more frequently, such as whenever virus is detected by a viral load test," commented lead researcher Neil Parkin, senior scientist at ViroLogic. "Additional studies will provide us with more informed insight into the clinical and economic impact of performing phenotypic resistance testing at detectable low viral loads."
For more information: Sidney Ho, Director of Public Affairs, ViroLogic, 270 E. Grand Ave., South San Francisco, CA 94080. Tel: 650-635-1100. Ext. 7406, Email: sho@virologic.com.
Edited by Laura DeFrancesco
Managing Editor, Bioresearch Online
ldefrancesco@vertical.net
