Transposon-Tagging Goes High Throughput
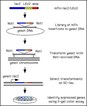
Using their transposon-tagging technique, the group has amassed several insertion libraries with different reporters and tags. All libraries and the reagents used to construct them are freely available, as are the data derived from them. Information on the libraries can be found on the Yale Genome Analysis Center web site (http://ycmi.med.yale.edu/YGAC/home.html).
Back in 1994, this group first reported on their technique for massively tagging individual yeast genes with mini-transposons containing mutagenized genomic DNA fragments fused with the b-galactosidase gene (Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. "Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae," Genes Dev 8(9):1087-105, 1994). In this work, 2,800 transformants were isolated that expressed fusion proteins, and when analyzed by indirect immunofluorescence, several hundred fusion proteins could be localized to discrete areas of the cell (nucleus, mitochondria, endoplasmic reticulum, cytoplasmic dots, spindle pole body, and microtubules).
In the present work, full-length epitope tagged versions of the genes were used in the library construction, a change that has increased the ability to localize the fusion proteins through immunofluorescence. In addition, the researchers have incorporated technology adapted from large-scale sequencing projects. By utilizing this technology, rather than using a shotgun approach for creating transformants, they can now work with individual bacterial clones, each containing an insertion-library plasmid, which are isolated and transformed individually into yeast. This has enabled them to scale up their effort such that screening thousands of transformants per week is possible. Once productive tranformants are identified—which is happening at the rate of 120 per screen—they can then return to the original plasmid to identify the genomic location of the DNA involved in the fusion, rather having to rescue the plasmid from transformants.

As more and more sequence data piles up, new technologies like this one will facilitate making the leap from sequence to function. In fact, in this study the researchers compared their results on identifying sporulation genes with those obtained through expression array analysis, and found that they turned up almost twice as many genes using their screen (31 by transposon-tagging versus 17 by microarray analysis). What this means is that this technique can not only identify previously unidentified genes, but also may prove to be more sensitive for expression studies.
For more information: Michael Snyder, Professor and Chairman of Molecular Cell and Developmental Biology, Yale University, P.O. Box 208103, New Haven, CT 06520-8103. Tel: 203-432-6139. Fax: 203-432-6161. Email: michael.snyder@yale.edu.
By Laura DeFrancesco
